Given the following JSON:
{
"Available online": "29 January 2020",
“Received": "8 November 2019",
"Revised": [
"16 January 2020"
],
"Accepted": "23 January 2020",
"Publication date": "1 May 2020"
}
The Date can be captured with dd MMMM yyyy:

Notes about using Pentaho Data Integration and Apache Hop as Non-Programmer
Given the following JSON:
{
"Available online": "29 January 2020",
“Received": "8 November 2019",
"Revised": [
"16 January 2020"
],
"Accepted": "23 January 2020",
"Publication date": "1 May 2020"
}
The Date can be captured with dd MMMM yyyy:

To get a first impression about the content and size I was using the Crossref Rest API facet search:

So for 2020 there are 542k entries. In order to download metadata data (or in my case just the DOI, date of DOI registration and the Elsevier internal ID) I’m using again the REST API of Crossref.
Actually a Crossref query is limited to provide max 1000 DOIs. Using cursors, it’s however to possible to loop further and get about 100k DOIs before the API times out. So in order to get all publications from 2020 I created a monthly batch based on the created date like:
https://api.crossref.org/works?filter=member:78,from-created-date:2020-01-01,until-created-date:2020-01-31&select=DOI,created,alternative-id,&rows=1000&cursor=*
In PDI I’ve created a job, that handles the cursor and repeats the transformation with the REST Query as long there is a new cursor coming back from Crossref.

So I get a list with all alternative IDs (like: S0960982219315106) from Elsevier articles. With that I can create an URL to all abstract pages of this article: https://www.sciencedirect.com/science/article/abs/pii/S0960982219315106
Using the Pentaho HTTP Client, it’s now possible to get the HTML of the this abstract page.

It’s important that there’s a HTTP-Header like: “Mozilla/5.0 (Macintosh; Intel Mac OS X 10_15_5)”, otherwise the Pentaho Client will be routed to a error page.

Elsevier makes it very easy to extract information about the article without going deep into the HTML-Structure of ScienceDirect. Within the HTML there’s a whole section with structured data in JSON:

Loading the whole HTML in one field, the following Regex can be used to extract the json block:
*<script type="application/json" data-iso-key="_0">(.*)</script> <iframe style="display: none.*

Having the JSON data block separated from the rest of the HTML, we now can pass it to the JSON-input step. There is really plenty of information and much more than you would see via front end.

For the moment I’m mostly interested in the author and affiliation data:

In the JSON we can extract:

extracting the authors works similar like the affiliation.

An author can have multiple labels/refids, emails, contributor-roles. In order to get those flat in a row separated by a coma, a group step can be used, while getting it back into the stream using the “stream lookup”-step:

In a third step authors and affiliations are merged by the the label:

in order to get a list with authors and affiliations

You can bundle a couple of steps as a transformation and call those steps in another transformation.
I often use the Crossref Rest API to get information about publications. Depending on the publisher there are different kind of dates associated with a DOI and the dates can have different resolutions. Sometimes just a year or a year and a month.

In order to get always a specific publication date with the resolution YYYY.mm.dd I use a couple of steps and logic to determine the “relevant” publication date out from those different date fields.

To reuse those steps in different transformations without copying each time all these steps I can now save those steps as own transformation. Let’s add a “Get rows from result” and “Copy rows to result” at the beginning and and end this sub-transformation.

Then we can add a “Transformation executor” step in the main transformation. In this step we add the expected “fields” of the sub-transformation in the tab “Results row”
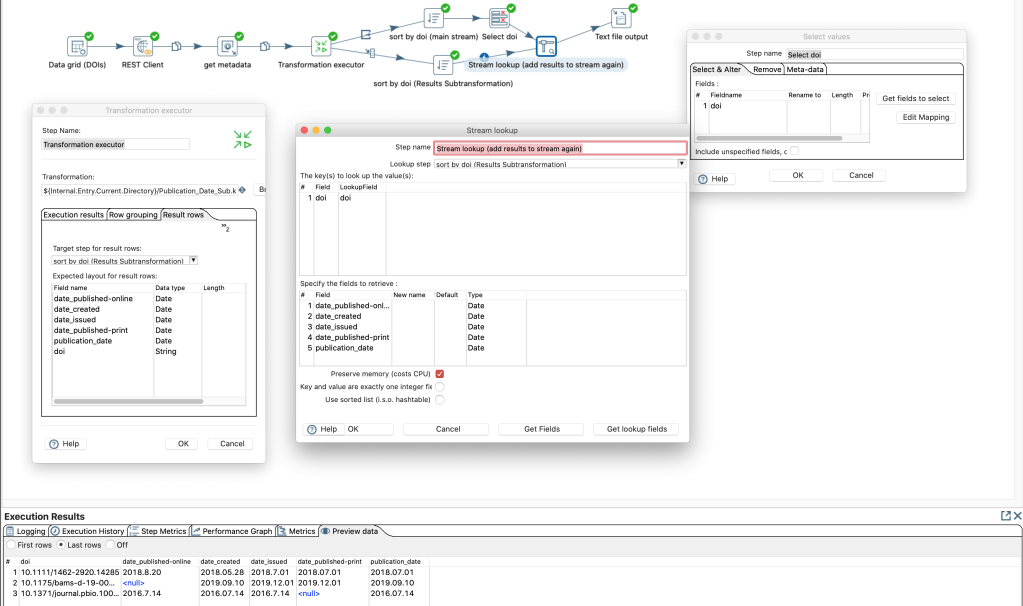
As output of a “transformation executor” step there are several options available:

There seems to be no option to get the results and pass through the input steps data for the same rows. Probably since the output of the sub-transformation can have more more or less rows than the input. Yet we can create a work-around by going on the the input data and add the results of the sub-transformation with a common (presorted) identifier. At the end we have the original data and the result of the sub-transformation combined.
Pentaho Data Integration (PDI) offers the input step “JSON-Input” to read out data from a JSON file or stream. Often I use this step after a REST-API-Query, so I would have the JSON-Input as a field from a previous step.
In order to test the field-extraction, it’s helpful to save some local samples of the possible responses. In the tab “File” you first can do your tests with the local file and switch later to “Source is from a previous step”.

Since recently Pentaho offers an “internal helper” to select the fields. However it unfortunately does not to work for most of my use-cases. Instead I found http://jsonpathfinder.com/ very useful.

Then add the fields with the corresponding path in PDI:
