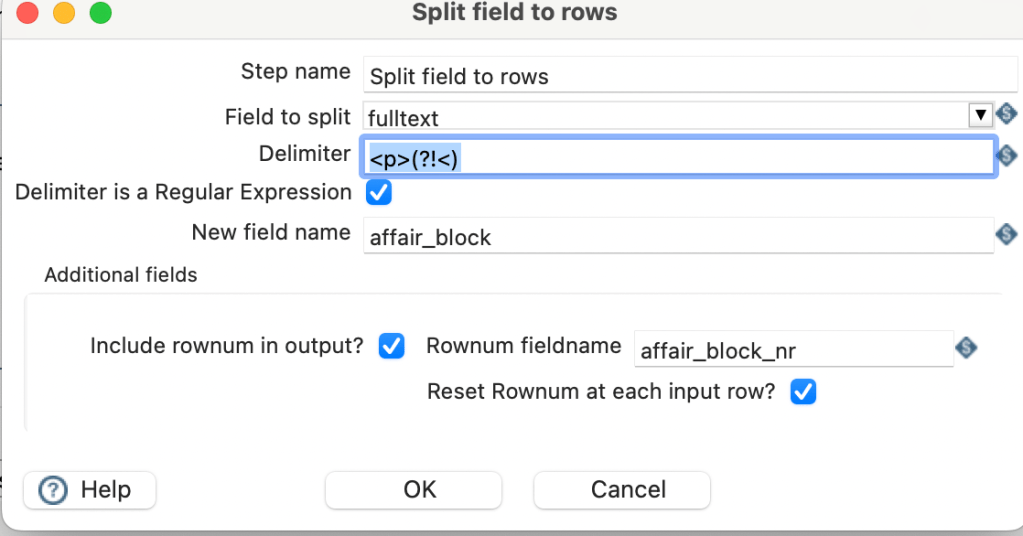
Currently PDI and Apache Hop do not run natively on M1. There are reports using the ARM optimised Azul JDK, but this was not working for me, as the newest STW Library is not yet ARM compatible.
Instead the following procedure worked for me on my Apple Macbook M1 (1st Gen).
Installation JDK 8.
If other JVMs are installed, they should be selected as follows:
Which Java versions do I have installed?
/usr/libexec/java_home -V
results in my case Matching Java Virtual Machines (3):
16.0.2 (x86_64) "Oracle Corporation" - "Java SE 16.0.2" /Library/Java/JavaVirtualMachines/jdk-16.0.2.jdk/Contents/Home
1.8.0_302 (arm64) "Azul Systems, Inc." - "Zulu 8.56.0.23" /Library/Java/JavaVirtualMachines/zulu-8.jdk/Contents/Home
1.8.0_301 (x86_64) "Oracle Corporation" - "Java SE 8" /Library/Java/JavaVirtualMachines/jdk1.8.0_301.jdk/Contents/Home
Set Java_HOME Path correctly:
unset JAVA_HOME
export JAVA_HOME=$(/usr/libexec/java_home -v "1.8.0_301")
Check selected JVM
java -version
echo $JAVA_HOME
Set terminal to Intel mode according to: https://stackoverflow.com/questions/67972804/pentaho-data-integration-not-starting-on-new-mac-m1
And replace SWT.jar library (according to previous step)
With that changes, you can start PDI and Apache Hop on M1